Building cloud-native solutions is not only about adopting new architecture paradigms but also embracing a new mindset around agile, distributed DevOps. At CSIRO's Transformational Bioinformatics group we demonstrated how converting traditional software tools to cloud-native solutions can save resources and open up new application areas.
Starting our cloud-journey in 2016, we built one of the first research applications using entirely serverless technology. Since then we have made a sport out of discovering how to convert traditional widely used bioinformatics tools to serverless solutions or, as Computerworld puts it, "strangle [bioinformatics] monoliths".
What exactly is Serverless?
The Serverless or Function-as-a-Service model aims to abstract away everything but the code you write, thereby offering a consumption-based cost model and applying the economy of scale to orchestrating efficient compute.
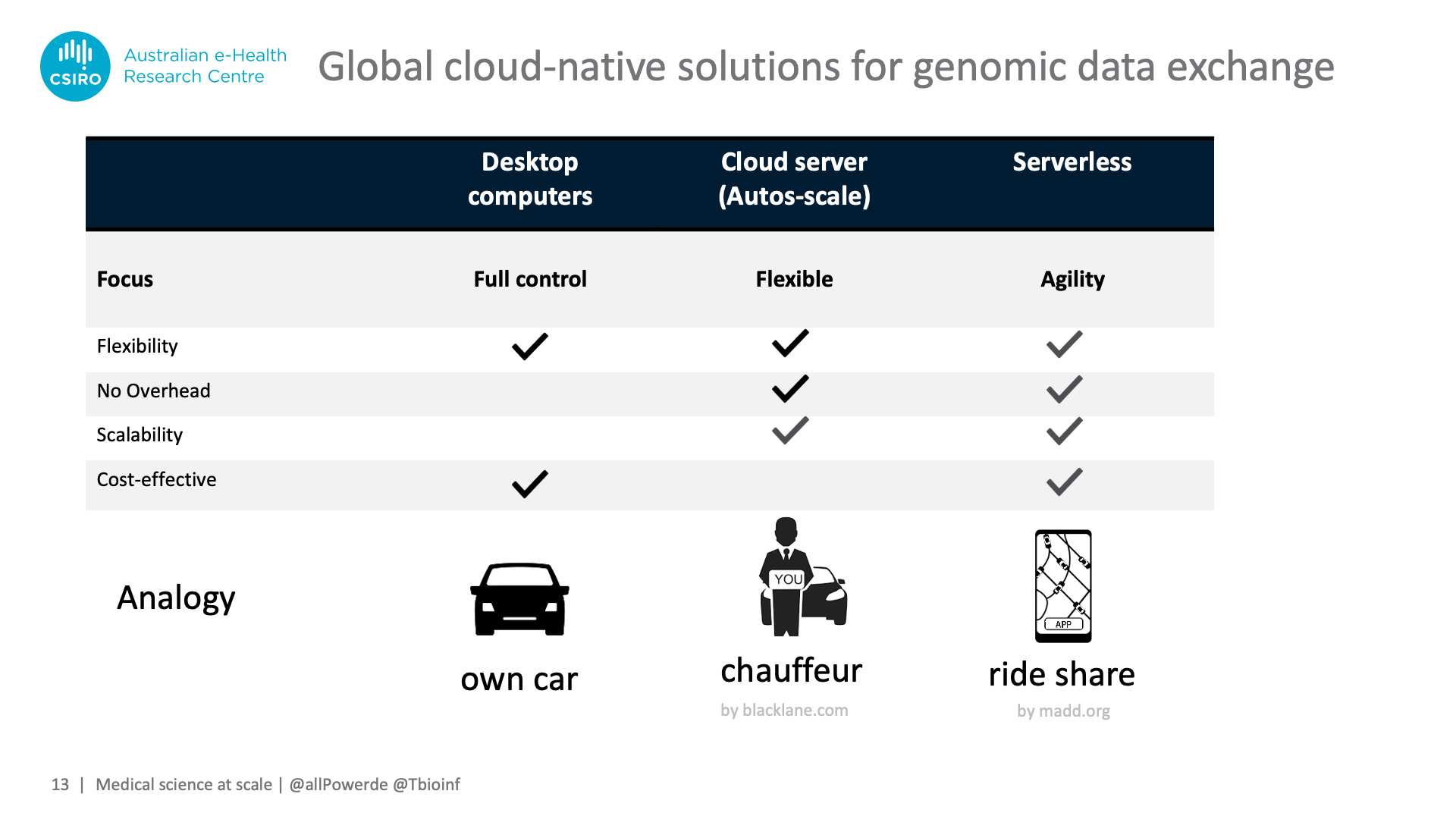
However, both "serverless" and "functions" are misnomers and can cause fierce debates when taken literally, so we like to use the analogy of ride sharing apps to describe the concept: Owning a car enables you to go wherever you want, however, you have to service this car and cannot easily upgrade to a bigger one if needed – our analogy for on-premises desktops or servers.
Ooh, “ride share” as analogy for serverless. I like that! Might have to retire my usual “takeaway food = kitchenless” example. @allPowerde #cloudstrategy19 pic.twitter.com/PiyW0r9pe2
— Kris Howard ? #BuildStuffLT (@web_goddess) September 26, 2019
You can off-load servicing chores and gain flexibility if you hire a chauffeur and own multiple cars, however this comes with a price tag – equivalent to spinning up beefy virtual machines in the cloud and auto-scaling them to different workloads.
Serverless combines servicing-freedom and scalability with cost-effectiveness. Like ride-sharing drivers having become so ubiquitous that being assigned the right type of car is virtually instantaneous, serverless can automatically "recruit" the right amount of CPU time or facilitate access to data without lengthy spin-up times for virtual machines.
This efficiency-boost comes with the downside of giving up a level of control, e.g. your tasks have to fit in within relatively rigid runtime and memory limits. However, like ride-sharing companies needing to stay on your good side offering tracking and rating systems, public cloud providers work on giving control back, such as allowing you to choose the language for the function, the execution memory size, and how to shield your application from other workloads on the same server. For example, AWS Lambda summarised its journey over the last 4 year since release, with "tl;dr: its a lot [of updates]".
4 years since the initial #AWSLambda release and us building @gt_scan2 as one of the first serverless applications in research. We have come a long way since then with @gt_scan2 more accurate and resource efficient as well as supporting a whole range of #genomeediting use cases. https://t.co/R3pu80L4AW
— Denis Bauer (@allPowerde) November 15, 2019
Serverless is adopted by all major cloud providers
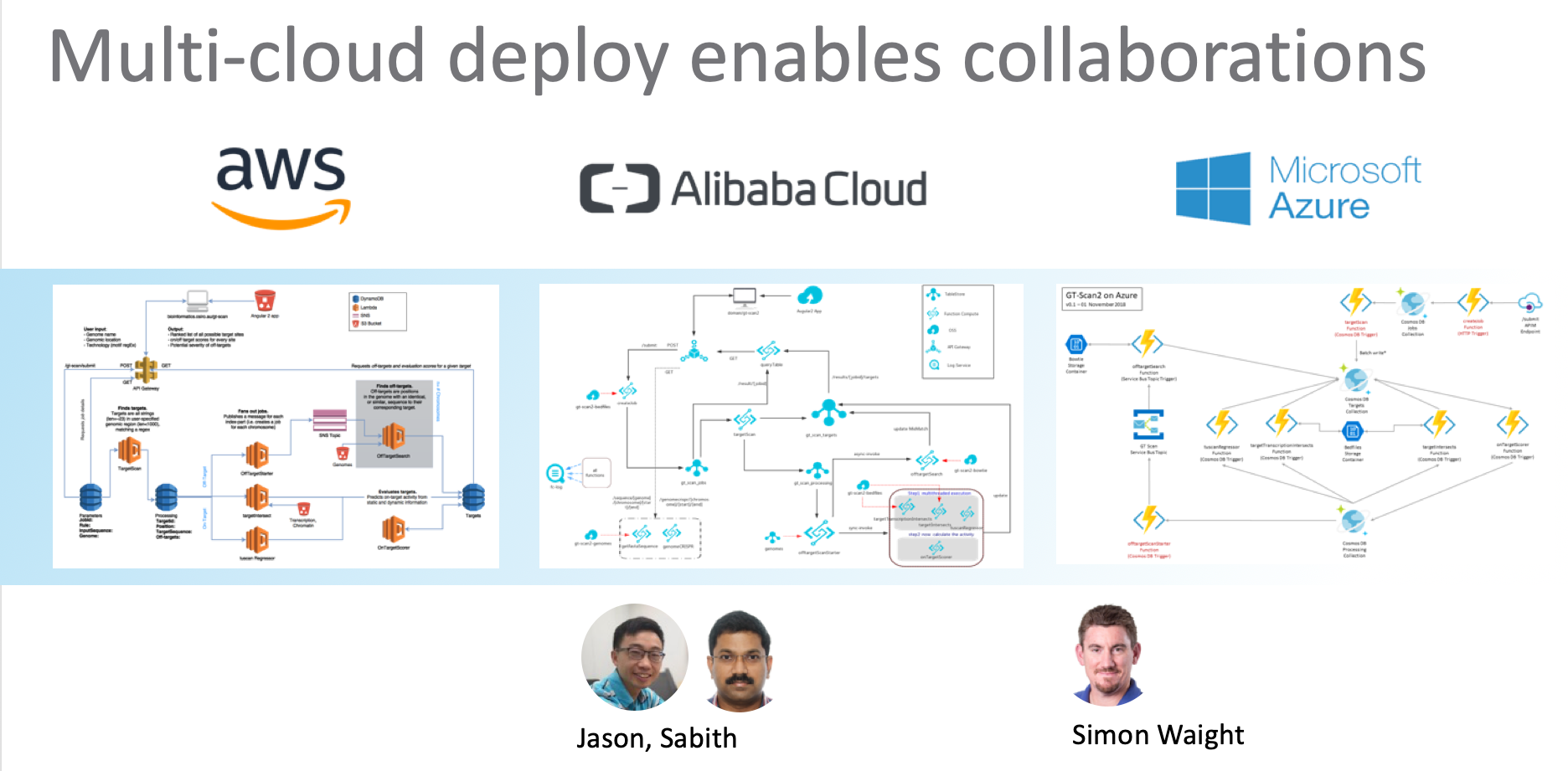
We developed the GT-Scan Suite on AWS, but it has since been re-implemented for Alibaba Cloud and Microsoft Azure. In fact, all major cloud providers and some of the smaller vendors have adopted serverless across the stack or at least for their runtime. For an overview see the Serverless Wikipedia page.
When does it make sense to use serverless?
We see three application areas where it makes sense to go serverless:
- Unpredictably fluctuating workloads or frequent low usage
- Processing large distributed data sources
- Building highly modular solutions potentially incorporating external services
We discuss these three points on recent architectures we developed.
Use case 1: serverless reduces cost by 99% for unpredictably fluctuating workloads (GT-Scan suite)
The GT-Scan Suite is a "search engine for the genome", where researchers can find the right editing spot for a wide range of genome engineering applications, including CRISPR. As such it was important for this search engine to be a web-based offering and "always on".
However, with the genome having 3 billion letters, finding the one efficient and unique site for the editing machinery to land on can be a computationally expensive task. As such, the web app can command 100-1000 CPUs at peak time but can also sit completely idle for long periods of time when no researcher is accessing the service. In addition, reducing response times requires a cloud-based solution that offers the service for multiple geo-locations.
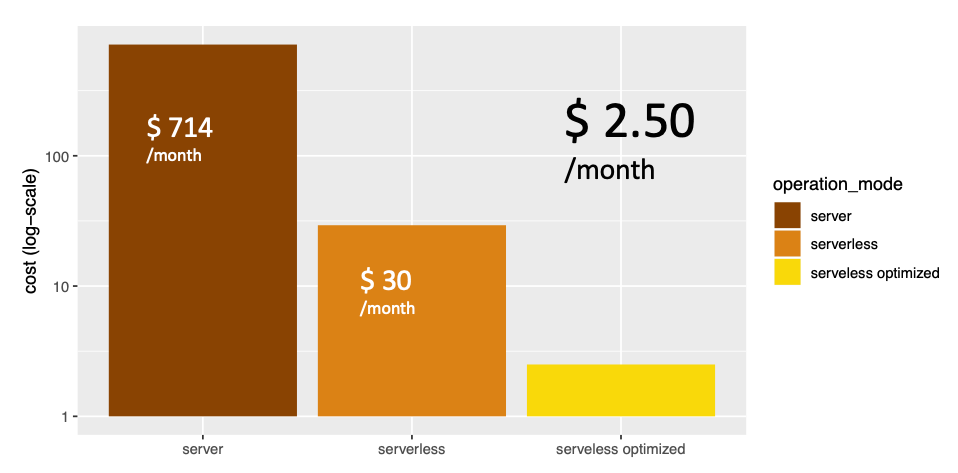
Proving such a cloud-based genome analysis service without initial wait-time for on-demand queries would require a persisting machine that is beefy enough to handle the occasional medium to large search tasks. On AWS, that would be an EC2 instance costing $714/month.
Serverless allows us to reduce this cost by 95% by only paying for compute resources when they are actually used. In AWS that means when CPUs are triggered from a Lambda function or data is stored in the DynamoDB database. We can further reduce this by optimising our architecture using autoscaling on the DynamoDB database, bringing it down to just $2.50/month.
"Once you go serverless you never go back..." says @allPowerde, lead at CSIRO. See how her team used AWS Lambda to architect a genome search engine that handles bursty workloads. https://t.co/dzdVVc7XqQ pic.twitter.com/skFUmuVpyt
— Amazon Web Services (@awscloud) November 12, 2019
Use case 2: processing large volumes of distributed data 5000-fold faster with serverless (sBeacon)
Finding disease genes is a daunting task as there are on average 2 million differences between any two people. Differentiating disease-causing mutations from our normal genetic differences requires information from as many genomes world-wide as possible. Sharing this data securely and in a privacy preserving way can be done with the Beacon protocol [1].
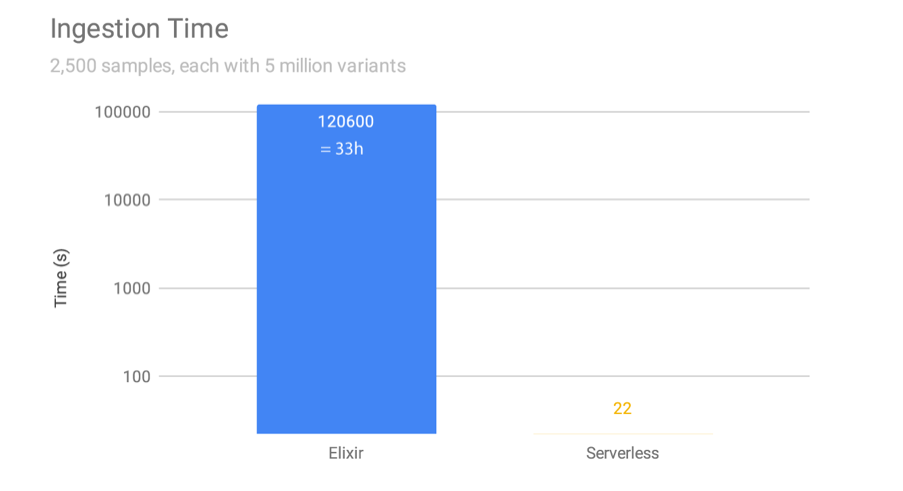
However, the traditional implementation requires all data in one Beacon to be consolidated into one enormous database before information can be served out to clinicians and researchers. Ingesting new genomes into the database on an AWS EC2 instance can take 33 hours, which for a daily intake schedule is not sustainable.
We re-implemented the Beacon protocol on serverless architecture: sBeacon allows people's genomes to be kept as separate flat files on object storage services, such as AWS S3, which reduces ingestion time from 33h to 22 seconds (5000-fold).
Furthermore, in sBeacon the subsequent queries can draft parallel functions (e.g. Lambda) to collect information from the relevant sections of the files only, which reduces costs (from US $4000 to US $15.60) and enables the run time to be kept constant (20 seconds down to 1) as the load increases (with 100,000 individuals and 85,000,000 variants). It also aids in the application of differential privacy and helps preserve data ownership.
Use case 3: Building modular future-proof applications (sVEP)
Genomic information is used in clinical decision making as 99% of patients have a genetic variation that affects clinical care [2]. For example, knowing adverse drug reactions in advance or determining a specialised treatment pathway can dramatically improve patient outcomes. Curating the correct information out of the wealth of available medical and scientific knowledge in order to annotate a patient's mutation has become an art form practised by clinicians and pathology labs alike.
Hence, being able to tailor a decision-tree-like workflow to selectively add bespoke annotations while skipping irrelevant information is vital to maintain clinically relevant turn-around-times as more and more information and methodologies become available.
Re-implementing the popular Variant Effect Predictor (VEP) using serverless architecture, sVEP is highly modular hence allowing different workflows to be processed efficiently by only consuming the resources needed. This allows users to define and re-define workflows on the fly, adding new annotation modules without disrupting the underlying architecture.
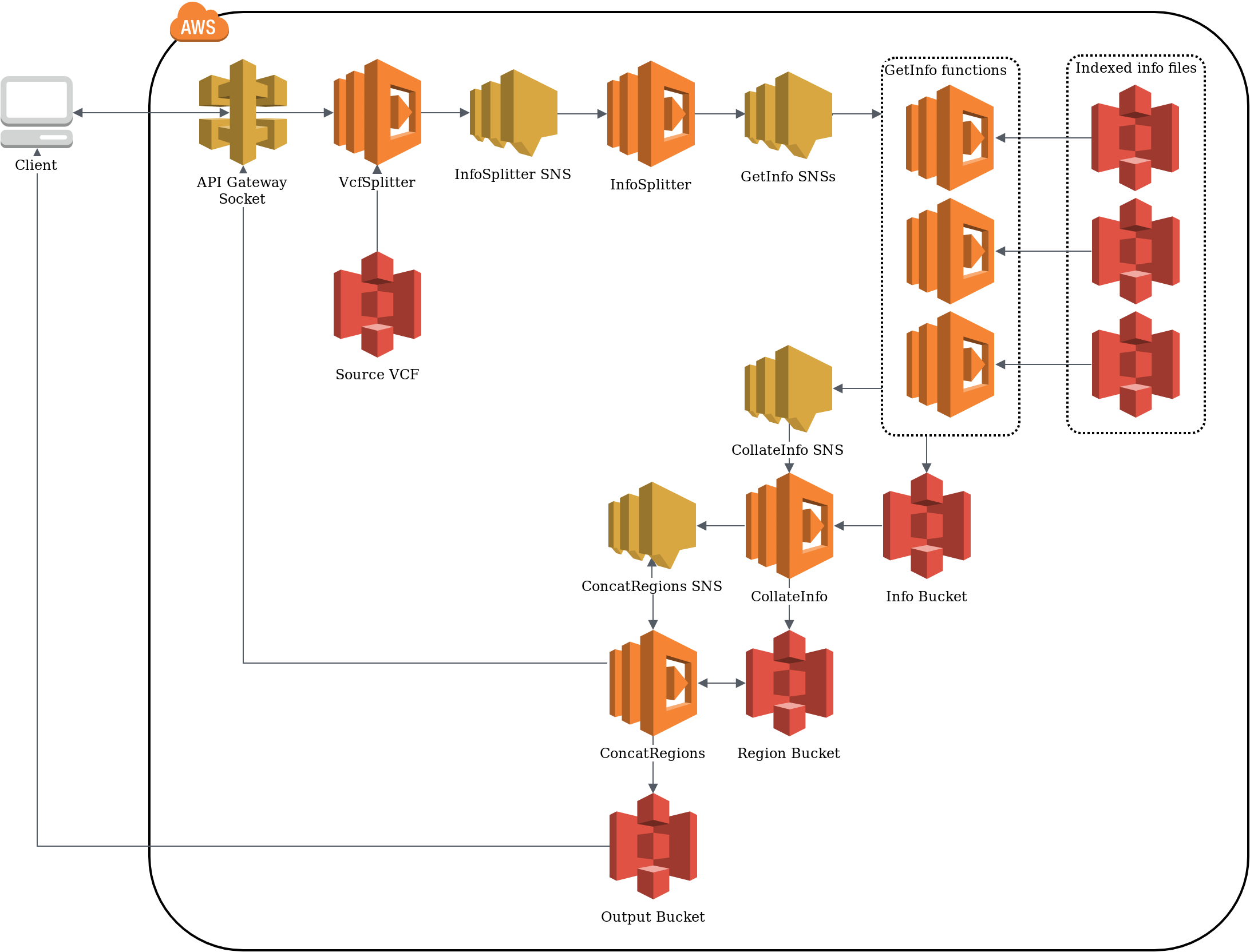
Developing optimal serverless architecture is not trivial
Using distributed, modular services makes optimising the architecture difficult. By now, cloud providers offer several hundred services with very nuanced differences. This makes it impossible to choose the optimal architecture up-front without experimentally testing the setup to record run time and resource consumption.
In our DevOps.com article, we advocate for a hypothesis driven evolution of architectures. Here, two identical architectures are deployed side by side, with one representing the current state of the architecture, and the other containing the variation that should be tested empirically. This allows us to answer questions such as how much can be saved by switching a dynamoDB to on-demand capacity.
Our approach of applying the scientific method to software engineering has been published by @devopsdotcom "DevOps 2.0: A New Evolution of Serverless Cloud Architecture" - https://t.co/BewYag9pLh https://t.co/B9Ee9z0Mgb https://t.co/Ov0SpvhNhU
— Denis Bauer (@allPowerde) November 18, 2019
Besides optimisation, maintaining serverless architecture in a continuous deployment procedure requires some tweaks. Hear more from our lead developer at the 2019 YOW! Data conference:
Where to from here?
Are you interested in converting a traditional application into a serverless cloud-native solution? Do you want to team up with an experienced partner to get the architecture as resource and cost effective as possible?
Then get in touch to have a chat about how we can help.


