To fight COVID-19 we need to understand the functional consequences of viral mutation. However, only 13% of current entries in the world’s largest COVID-19 virus database (GISAID) have the necessary information about patient outcome. Together with GISAID and international researchers from Singapore and France, we have created a new data entry standard, published in https://onlinelibrary.wiley.com/doi/10.1111/tbed.13892.
What should be collected?
Nature journal stated “Unless each sequence of the virus in the global open repositories comes with additional patient information, the practical benefits of such record sequencing are lost”. Recognizing the need for clinical data, GISAID enabled 'patient status' to be recorded for each entry and made this field mandatory as of 27 April 2020. However, by 01 October 2020 only 13% of entries were anything other than "unknown".

While the WHO has recommended a “severity score” that collapses symptoms and outcome in a single score, this is likely insufficient due to the extreme diversity of outcomes and ongoing association of new symptoms. It is hence critical to collect the “patient journey” in as much detail as possible to understand the acute disease and potential chronic consequences.
What hampers the progress?
The lack of digital infrastructure for collecting clinical information has hampered progress. Here a standardised vocabulary and mechanism for linking in with the health system (interoperability) is needed to support health care professionals in capturing the necessary information in a way that does not divert time from their primary focus: saving lives.
CSIRO has developed such an infrastructure using the international standard Fast Healthcare Interoperability Resources (FHIR). The developed FHIR questionnaire is able to convert and capture free text into standard vocabulary and collects de-identified information about symptoms, vaccine status, travel history, and other relevant data that will help assess the individual patient journey and put context to the collected genome sequences. This resource is now published in Transboundary and Emerging Diseases as a collaboration with colleagues from Institute Pasteur, Singapore’s A*star and GISAID.
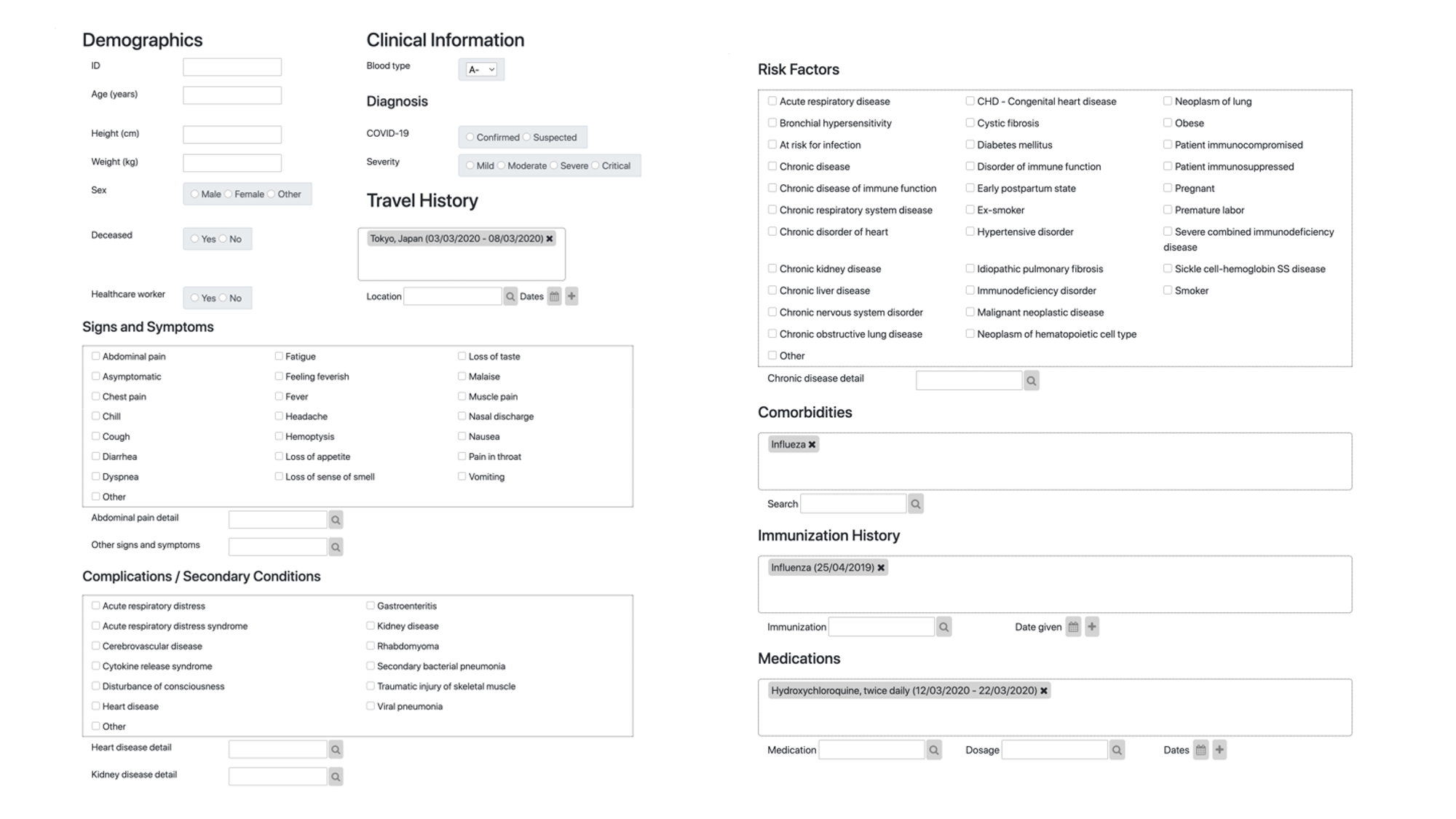
Road to adoption
The road to adoption is likely long as there are other non-technical roadblocks identified in the study, such as patient status being actual "unknown" to the submitting pathology lab. A consolidated effort between Pathology labs and clinical systems is hence needed, and FHIR-based data exchange is likely the key for enabling this.
Covid-19 mink variants discovered in humans in seven countries https://t.co/7kLTqNd8kB
— The Guardian (@guardian) November 18, 2020
Subscribe to Transformational Bioinformatics
Stay up-to-date with our progress


