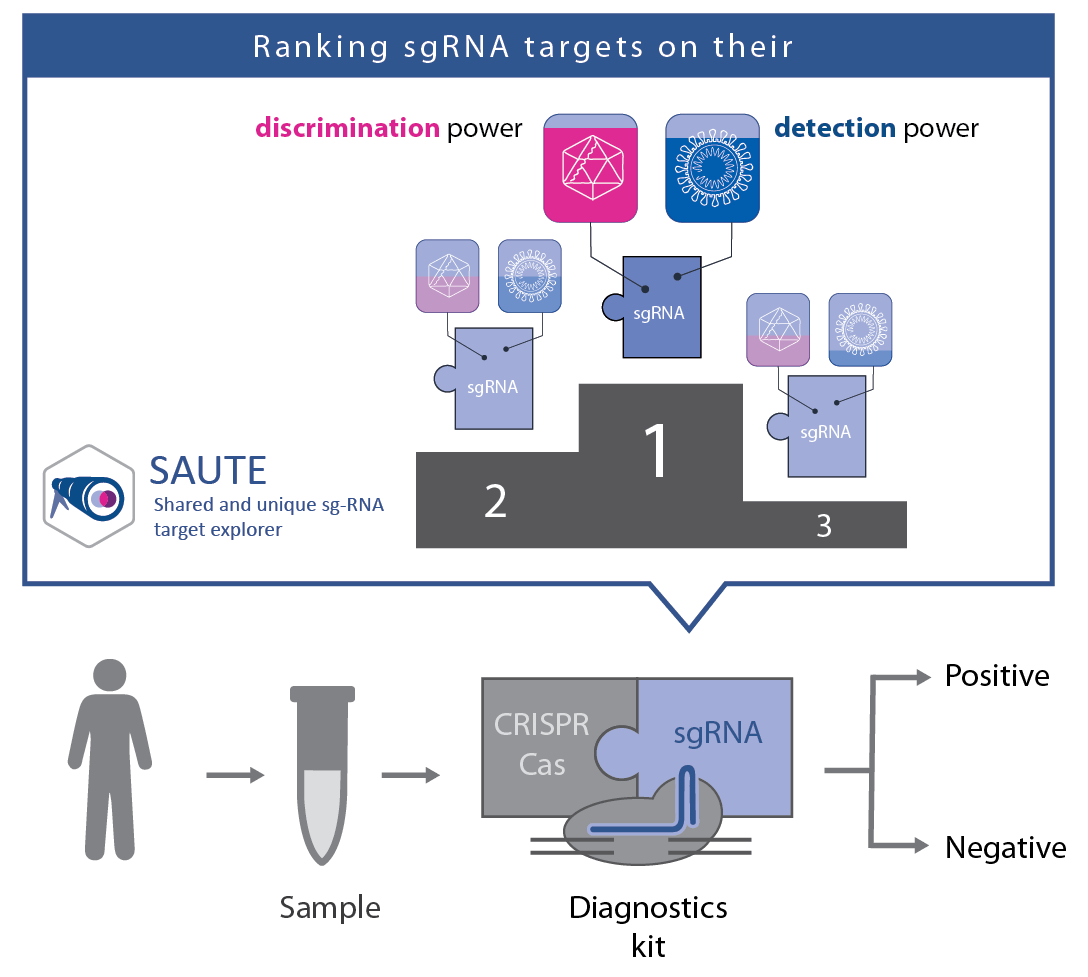
SAUTE (Shared and unique sg-RNA target explorer) is a tool to identify and rank sgRNA across multiple organisms. The user may select one or several organisms from which all common target sites will be found. The user may also specify the rule by which the targets will be identified, the default is the CRISPR-Cas9 System. Optionally, users may also enter organisms from which the candidate targets must not appear.
For added specificity, users may identify regions within the genomes for blacklisting or whitelisting from the search.
Output is grouped by organism, and the number of off-targets up to a specified amount of mismatches is provided for each target in each organism, to aid in identify optimal targets to fit the users use case. Output can be sorted as necessary.
Availability
- Custom implementation
- Bespoke solutions
- Product workshops
- Managed updates
- Full support

