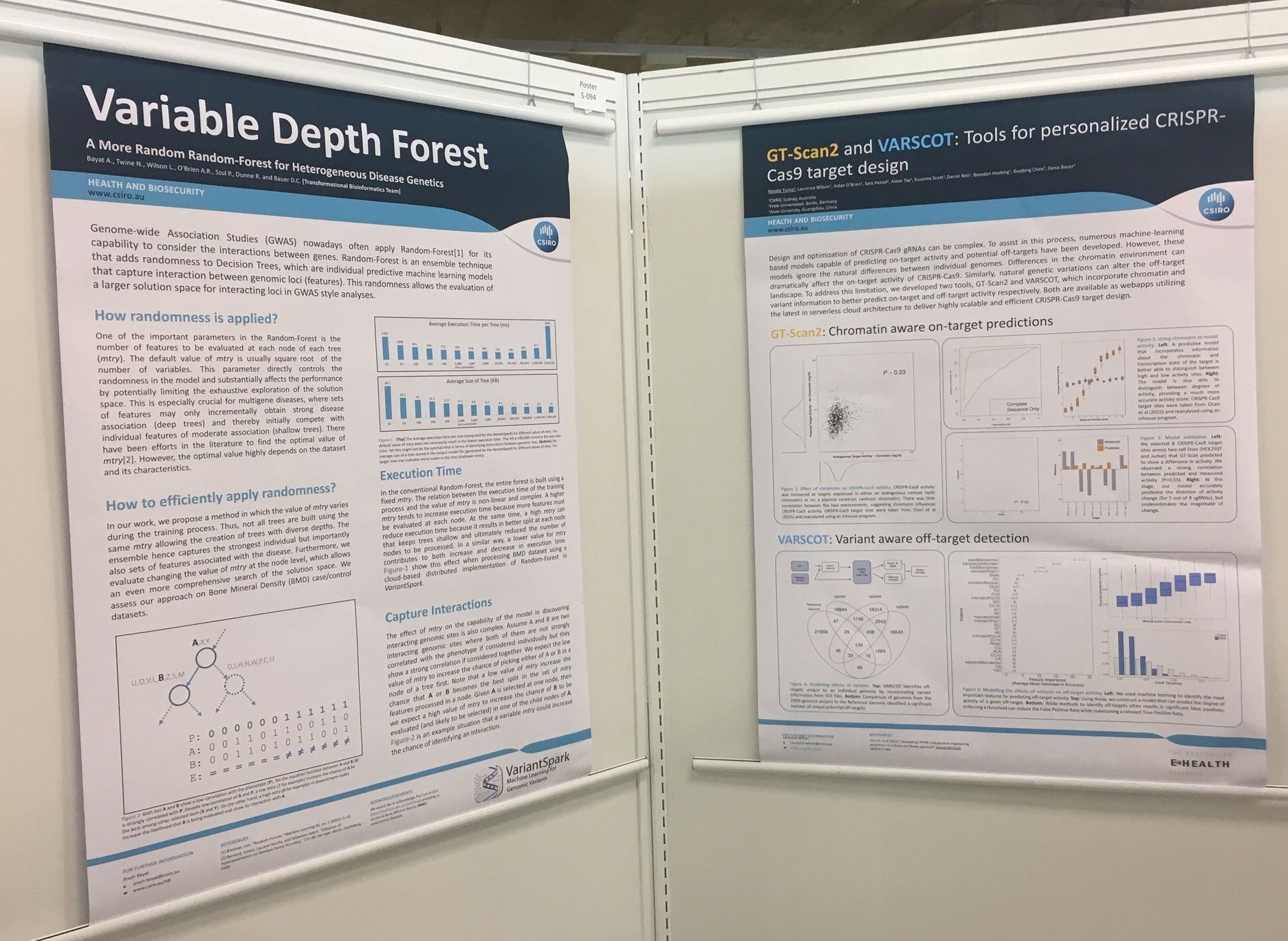
This July I was fortunate to present at the largest international bioinformatics conference, ISMB-ECCB. It was held over 5 days in Basel, Switzerland and attracted over 1000 attendees with 10 parallel sessions each day. Eminent Professors including Nikolaus Rajewsky, William Noble and Bonnie Berger delivered fascinating key notes covering topics from Deep Neural Nets, Single-cell RNA and Biomedical data analysis at scale. My presentation on 'TRIBES' received alot of interest, as did our posters on GTScan suite and VariantSpark. We also won 3rd place in the Terra competition!
Keynote speakers captivate...
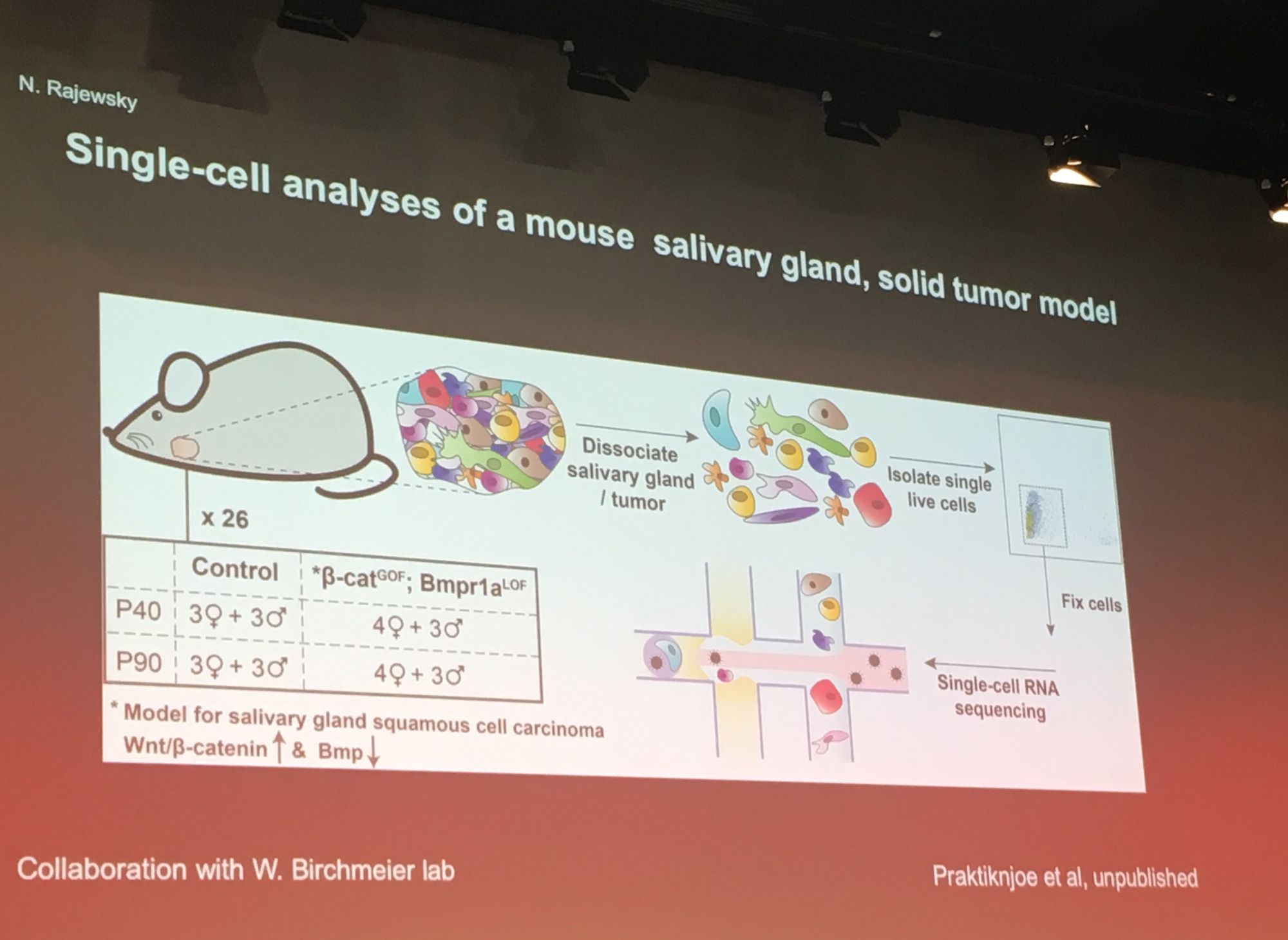
A popular theme throughout the conference was utilisation of large scale single-cell RNASeq data. Professor Nikolaus Rajewsky from MDC Berlin kicked off proceedings with his keynote exploring gene regulation in space and time using single-cell analysis. Professor Rajewsky's group generated a 'virtual embryo' by sequencing most of the cells in a fly embryo, with ~8000 genes per cell. Researchers can explore this resource at www.devtex.org. He also applied his methodology to reconstruct a mouse salivary tumor at single cell level and compare this to a healthy salivary gland, highlighting the cellular and genetic source of the tumor.

The 'Netflix challenge' for biology
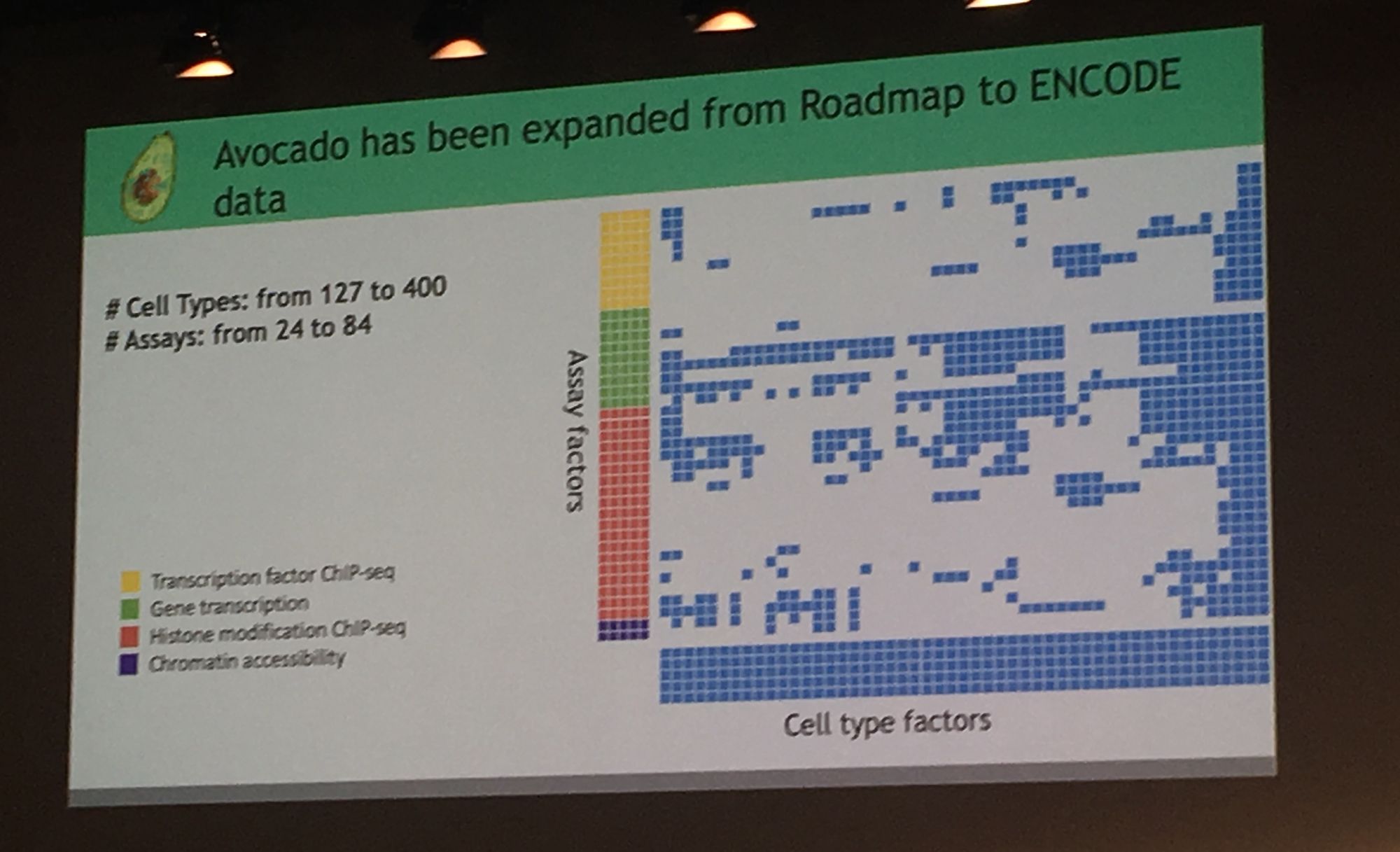
Professor William Stafford Noble from University of Washington continued the multi-dimensionality theme with his keynote: 'Travelling across spaces: the power of embedding genomic and proteomic data into a latent space'. Professor Noble's lab use machine learning approaches, specifically deep neural networks to explore 'omics data. His group are a fan of fruit names and Noble presented their 'Avocado' tool which uses a deep neural network tensor factorisation method to compresses genomic data from public repositories (e.g. ENCODE). Avocado can extract a low-dimensional, information-rich latent representation from experimental data. This representation allows the user to impute genome-wide epigenomics experiments that have not yet been performed. This was motivated by the 'netflix challenge' where one can impute missing data to predict shows a viewer may enjoy.

Data sharing and analysis at scale...
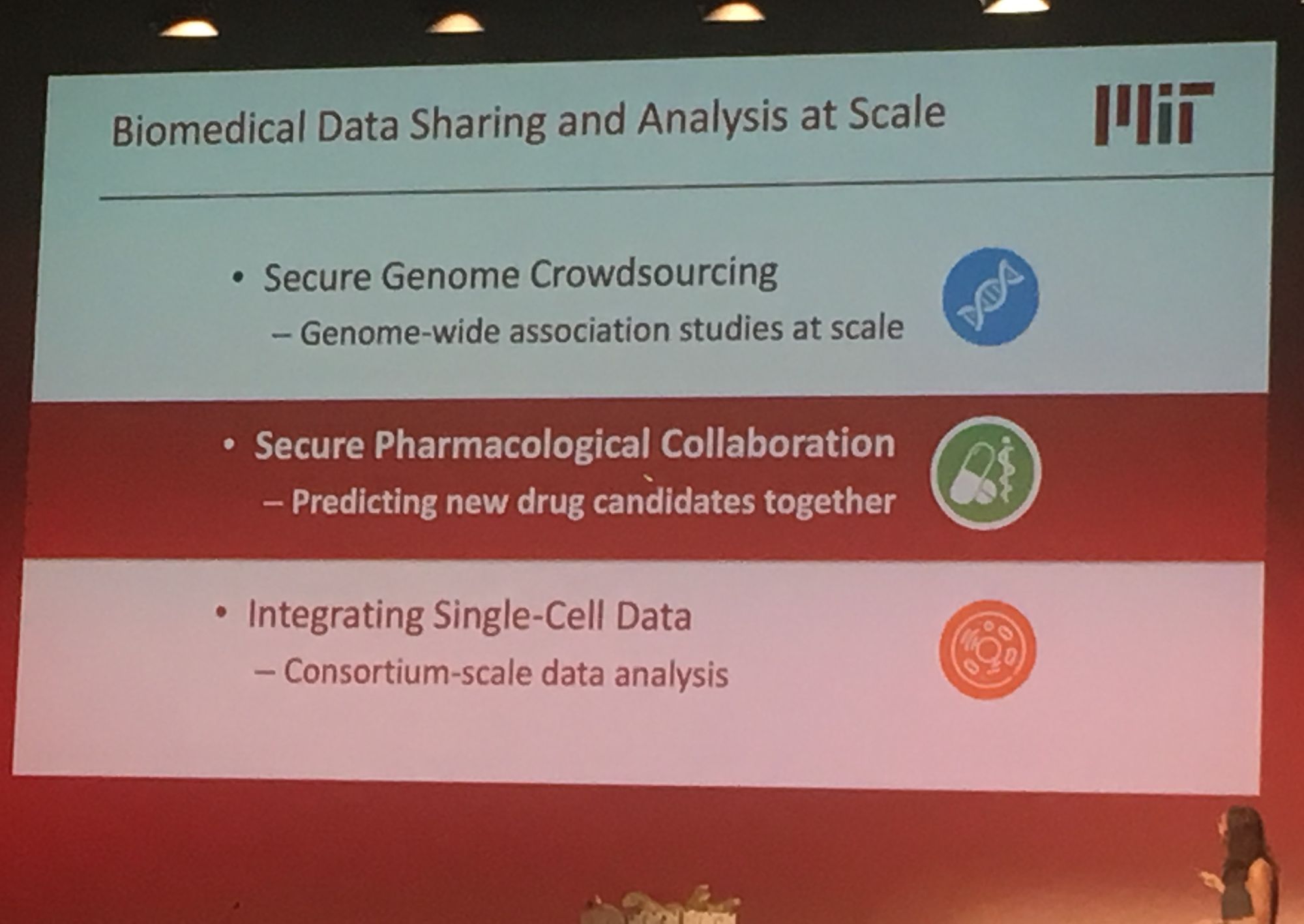
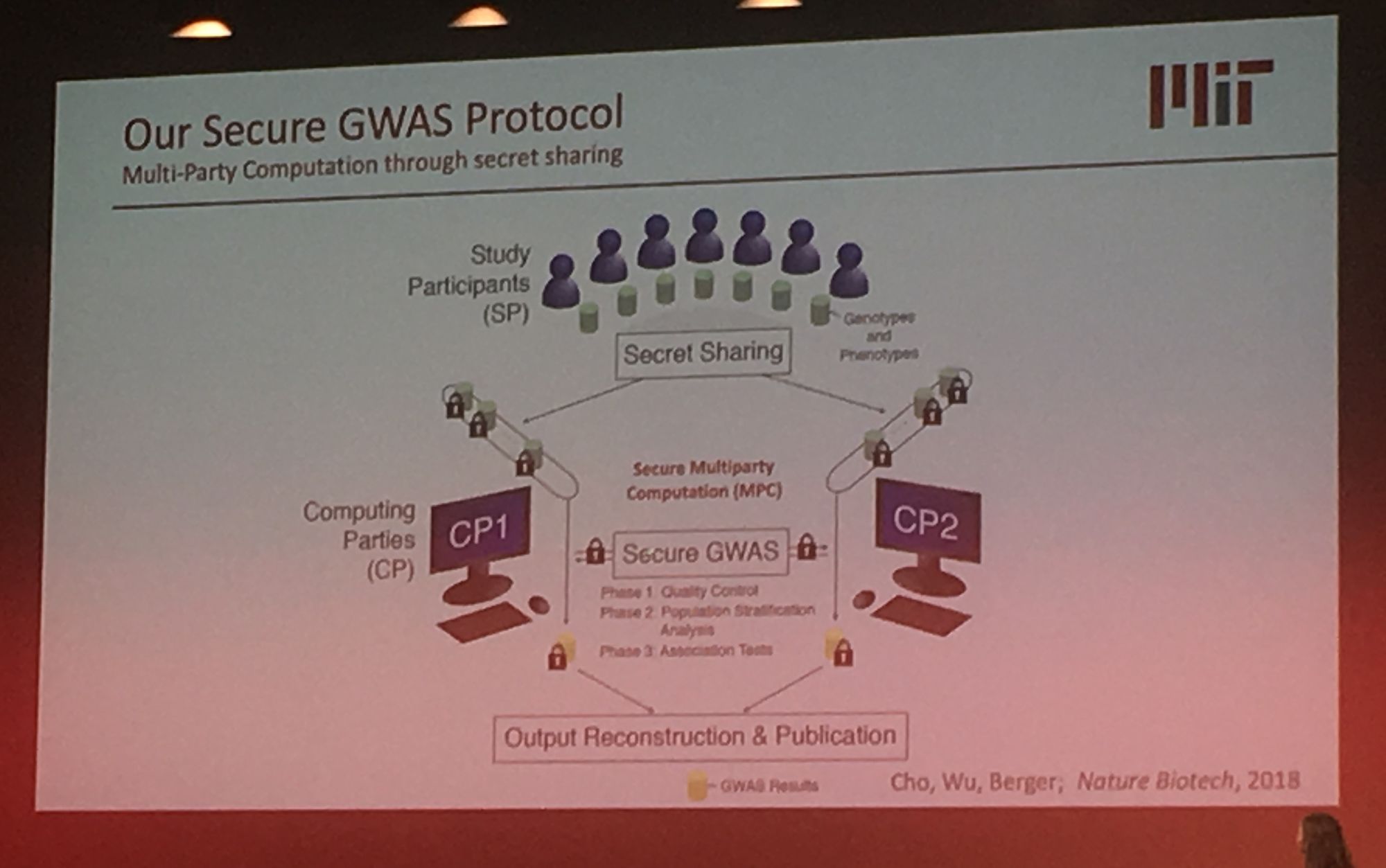
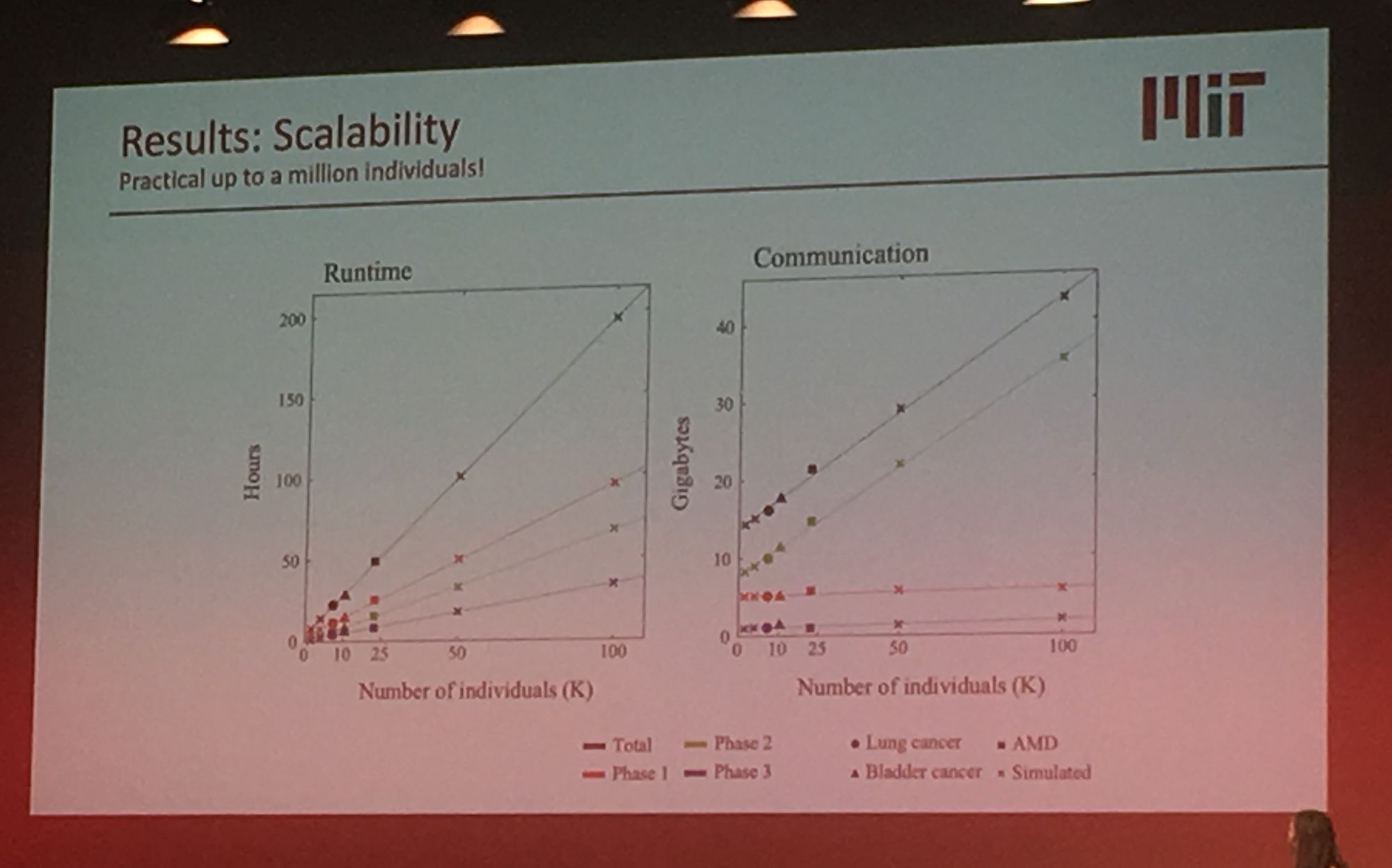
Sequencing data is growing exponentially and there is there is a pressing need for secure data sharing and analysis at scale. Professor Bonnie Berger from MIT addressed this in her talk: 'Biomedical Data Sharing and Analysis at Scale'. Prof. Berger addressed three key big data challenges for medical research: performing genome-wide association (GWAS) across multiple disparate datasets, in a secure way; Combining pharmacological data across the globe to predict new drug targets; and consortium-wide integrative single-cell data analysis. A topic highly relevant to our group's work is GWAS analysis at scale. Prof Berger presented their novel methodology which leverages secret sharing to enable secure multiparty computation. Here genomic data can be combined from multiple remote locations, in a privacy preserving manner, and GWAS statistics are then returned. Their secure-gwas protocol scales well and is practical up to a million individuals.
Our group was well represented...
I presented my talk on 'TRIBES' in the general Computational Biology section, this was well received and I had alot of interested parties coming to ask questions afterwards. We also had posters from the group on VariantSpark and GTScan Suite. And last, but not least, we won the conference 3rd prize in the Terra competition for our workflow on VariantSpark!
I look forward to more attending future ISMB-ECCB conferences!