The exchange of genomic and phenotypic data is becoming increasingly crucial by the day for delivering the best care for ailments with genetic root causes. Though the problem at hand may seem mundane on the surface, facilitating data exchange comes with a myriad of challenges. This blog post discusses the journey of building a serverless beacon, known as sBeacon, along with the associated design decisions and motivations.
The challenge
Sharing genomic data poses logistical and ethical challenges in the research space.
The Beacon protocol, developed by the Global Alliance for Genomics and Health (GA4GH), has emerged as the international standard to ensure that privacy and data ownership are preserved while enabling disease research. GA4GH protocols have been widely adopted by both the research community and adjacent industries. Although the protocol nearly meets the needs in facilitating research, its implementations lag in terms of completeness and scalability.
Existing reference implementations of Beacon are not scalable in terms of performance, resource utilisation, and, ultimately, the running costs associated with querying. Furthermore, their utility is further limited by a lack of certain aggregation functionalities, presumably due to technical and architectural design decisions. Hence, we have created a robust implementation, sBeacon, to address these existing limitations across the aforementioned criteria.
Developing sBeacon
We collaborated with the Australian Genomics Health Alliance and Genomics England to create a fully cloud-native architecture that enables the sharing of genomic data while preserving privacy and data ownership.
We moved past traditional computing and database management systems to a more modern, serverless architecture. Utilising AWS Lambda functions as the compute source, Beacon has been made into a fully serverless solution that can be automatically deployed in a user’s cloud account. The framework can run over patient-individual VCF files or large cohort VCF files. Owing to the parallelism of serverless computation, sBeacon can instantly scale to serve high query loads while being able to scale back to zero. This ensures zero compute costs during idle times.
Furthermore, we redesigned the data architecture by adopting the AWS Athena service. Athena is a serverless query engine for static files and supports SQL (which uses the Presto query engine). In contrast with reference implementations that used either MongoDB or PostgreSQL in older versions, Athena allows for scaling to zero and automatic scaling without committing to additional hardware resources.
Moreover, we designed sBeacon to query atop VCF files for genomic data. Firstly, this avoids an expensive step required to load VCFs into a database system such as PostgreSQL or MongoDB. Secondly, a VCF file is space-efficient, and a properly indexed VCF file can also be queried faster. In our experiments, we observed storing VCF files in MongoDB can be extremely space-heavy. Lastly, redundant storage of variants has many privacy and maintenance-related issues. For example, datasets can be taken down immediately from Athena, whereas removing millions of variant entries from a database system could involve inconsistent states or longer downtimes. Having just the VCF file can easily help users of sBeacon to onboard and drop datasets from sharing via the Beacon Protocol while they are being used for other research activities.
Overview of sBeacon
We evaluated sBeacon in a multifaceted fashion to investigate the effectiveness of our design decisions and the resulting architecture. We compared sBeacon against two other implementations of the GA4GH Beacon protocol version 2: Beacon RI (the reference implementation by EGA/GA4GH) and BSC Beacon (an implementation from the Barcelona Supercomputing Centre). We used the 1000 Genomes dataset, which includes 2504 samples, for this evaluation.
Features
Following is the complete visual summary of where sBeacon stands compared to the rest of the implementations.
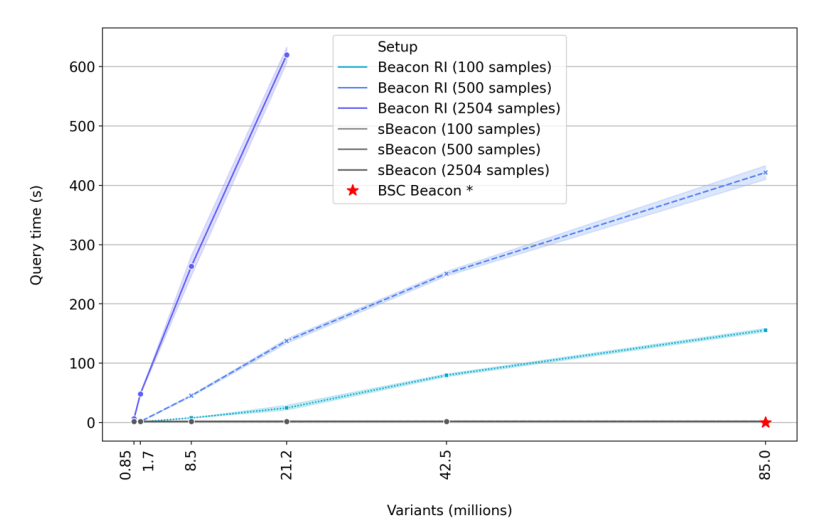
Query times
sBeacon manages to keep the query time to mere seconds, marking its exceptional efficiency.
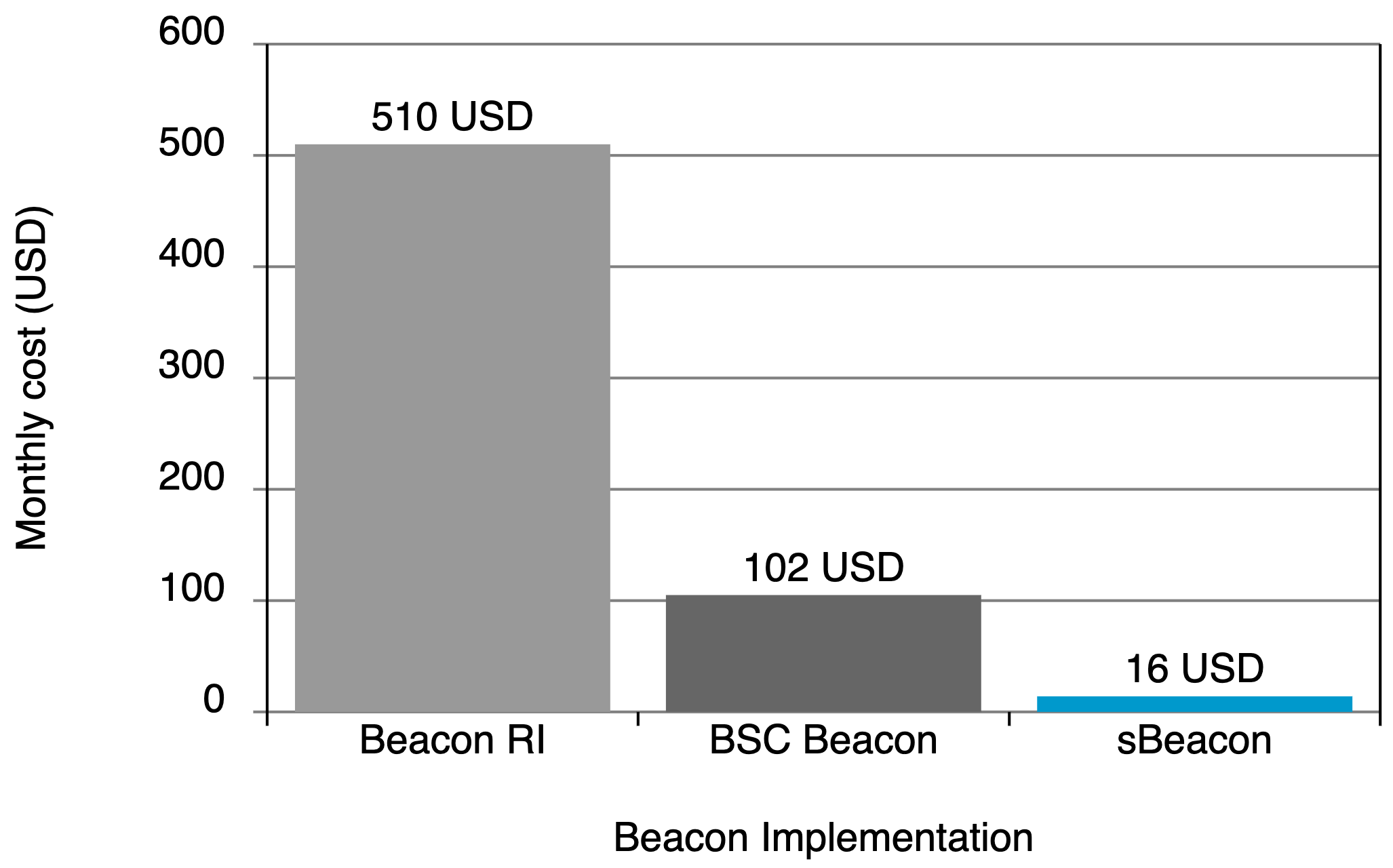
Running cost
While the competition can cost between US$100-500/month, sBeacon costs only around $16/month, even with an average of 72,000 queries per month.
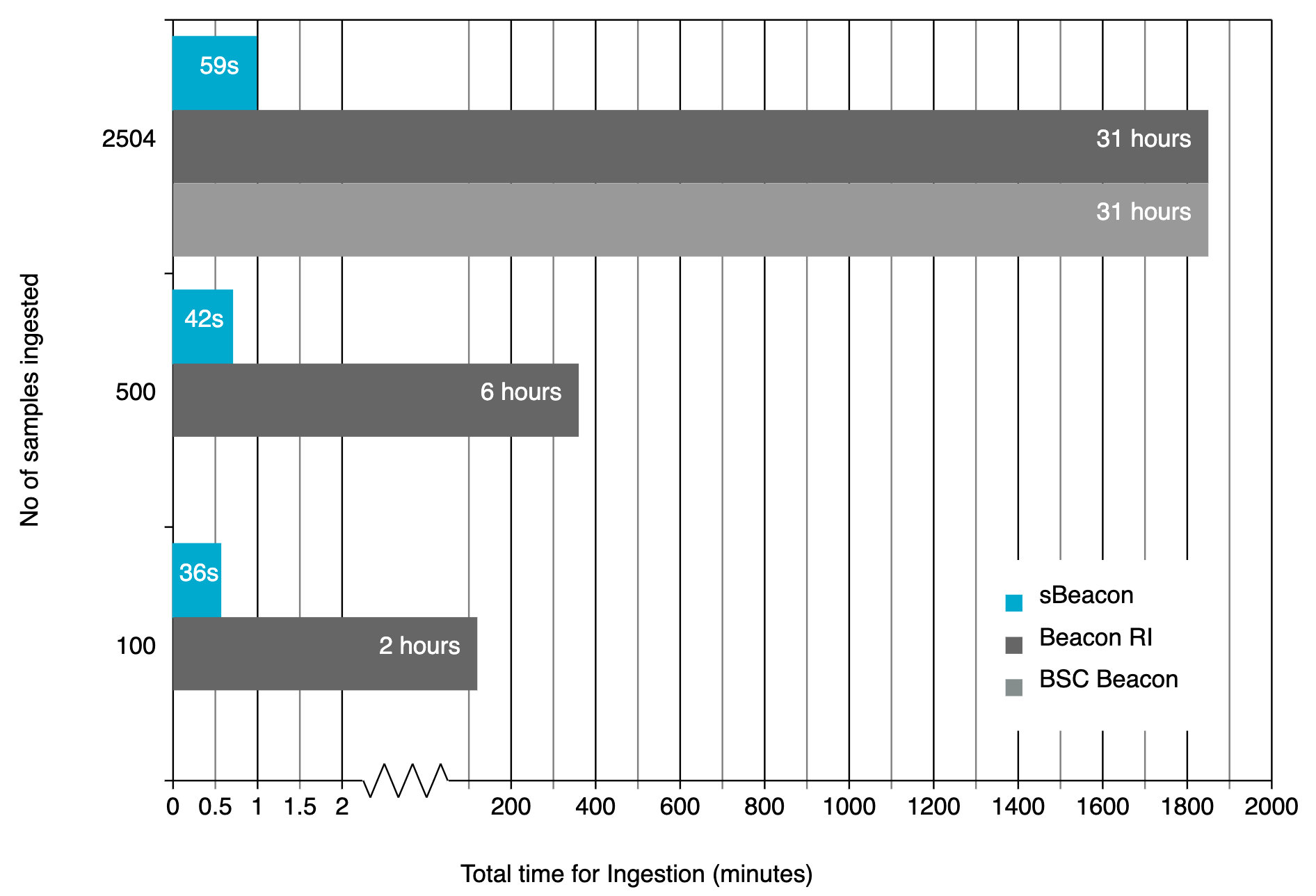
sBeacon does not transform VCF files, thus being able to ingest data with much lower onboarding times.
Conclusion
Developing sBeacon has been a great journey and it is our belief that it will set a positive trend towards more efficient and cost effective genomic data exchange.
Try sBeacon Today!
https://github.com/aehrc/terraform-aws-serverless-beacon/
References
If you want to work with the Transformational Bioinformatics group please visit Do Business With Us.
Subscribe to Transformational Bioinformatics
Stay up-to-date with our progress


