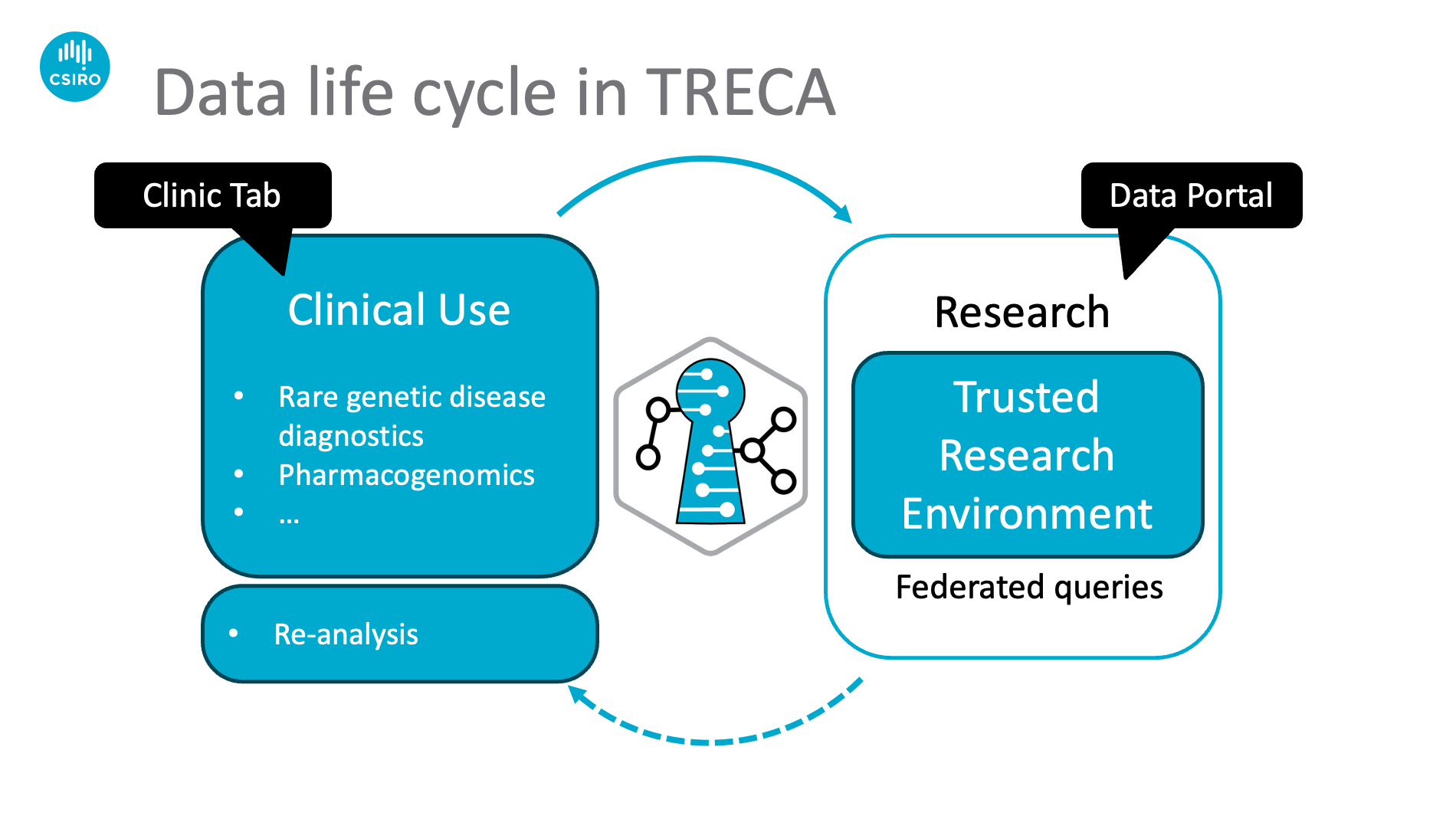
TRECA (Trusted Research Environment and Clinical applications) is an open-source cloud-based solution, supporting both clinical and research use of genomic data. It offers personalised diagnostics and medical decision support, before transitioning data into their research use within an air-tight digital vault to protect the sensitive and personally identifiable information and allow open-ended federated research.
While many genomic data management platform exists, they either serve clinical or research applications; not both. This is because either application has unique systems requirements that the other area does not need. Specifically, clinical-grade system require stringent quality control, robust auditable infrastructure and adherence to health privacy laws, while research requires a managed process for disseminating data, management of consent changes, and cater to ever-changing research workflows.

TRECA is designed to incorporate both elements as the boundaries between research and clinic are blurring with the advent of personalized treatments. Specifically, genomic data might be used beyond the medical application hence requiring even clinical applications to respond to a patients change in consent (dynamic consent). Likewise, increasingly stricter privacy laws will require even research systems to upgrade their security processes.
TRECA Clinic tab
TRECA's clinic portal can trigger highly customizable clinical workflows. For example, the rare-genetic disease workflow is powered by sVEP, our serverless implementation of EBI's variant effect predictor (VEP). This allows clinicians to efficiently and securely fetch annotations from databases and repositories around the world. Similarly, the pharmacogenomic workflow utilizes PGXFlow providing bespoke analytics on top of common pipelines such as PharmCAT. All modules are assembled to form a complete pipeline enabling clinicians to prioritize and annotate variants and generate a clinical PDF report.
TRECA Data Portal
Upon delivering the clinical outcome, the data will continue their life cycle inside the data portal. Here sBeacon, our serverless implementation of the Beacon protocol, facilitates querying genomic data. sBeacon also allows researchers to generate custom cohorts of phenotype of interest for population level analysis. Inside the TRE, TRECA enables unrestricted bioinformatics analysis through Jupyter notebooks and secure data exchanges within the platform. TRECA also empowers researchers with federated queries to genomic and phenomic datasets while also providing a Trusted Research Environment for downstream research after federated data discovery.
TRECA Security
TRECA is audited and closely monitored, maintaining human expertise throughout the process. Activities are logged, and resources are backed up to ensure sustainable and robust operation in a production environment. The platform uses cutting-edge cloud computing technologies to deliver cost effective and rapid analysis of data, adhering to data locality and strict access control demands.
Value Proposition
TRECA puts researchers and clinicians at the center of decision making freeing them from the constant concerns of data security, sensitivity and integrity.
Security: Ensuring the security of data and users is the key feature of TRECA:
- Trusted Research Environment - We created a secure virtual “vault” within which experimental and compute-intensive workloads can be executed.
- Role-Based Access Control (RBAC) - Personnel are onboarded within the system based on their specific roles, restricting unwanted application access.
- Authentication and authorization - Front-end and back-end is inaccessible without user authentication. Strong passwords are enforced within the system and MFA is strongly recommended.
- Web security - We implemented critical security elements like injection attacks prevention, cross site scripting, encrypted APIs, secure web headers, MFA for users, and no cookies usage.
- Audit trails - Every action within the system by any users can be audited upto 10 years. After 1 year the audits are archived (can be restored) for making the system cost efficient.
Real-world validation: TRECA is being tested in real-world via ministry of health in Indonesia.
Flexibility: With any research solution, flexibility is key to evolve the solution in different direction. TRECA is built with modular design and allows customisation for specific use-case.
ISO standards: TRECA is designed and developed in alignment with the principles of ISO 9001 standards, ensuring high-quality processes and best practices throughout its development
TRE in a box: TRECA is working out of the box through Infrastructure as Code deployment scripts.
Use Case: Biomedical and Genome Science Initiative
A TRECA instance is operated by the Indonesian Ministry of Health (MoH)’s Biomedical and Genome Science Initiative (BGSi) and delivered in collaboration with two Indonesian start-ups (GSILab and Xapiens).
- TRECA reduces the diagnostic odyssey for atherosclerotic cardiovascular disease (ASCVD) patients. The main therapeutic drug, Clopidogrel, has a 33% failure rate and non-responders are at risk of side-effects. With an estimated 500,000 Indonesians in this category and a 3% annual increase of ASCVD expected, this represents an growing cost to the health system. Using Pharmacogenomics (PGx) information, TRECA provides decision support to clinicians before treatment starts, achieving better health outcome.
- TRECA will enable the early detection of familial hypercholesterolemia (FH). An estimated 520,000 Indonesians are affected by FH, an inherited condition which has a high risk of developing Atherosclerotic Cardiovascular Disease (ASCVD), the most common cause of death in Indonesia named by the Health Minister, Budi Gunadi Sadikin. TRECA provides decision support to clinicians to identify FH early and is expected to reduce premature mortality and morbidity, avoiding approximately one cardiovascular death per ten FH patients screened (age 30–85).
- TRECA will increase rare genetic disease diagnostics. With at least 50% of rare disease cases still undiagnosed despite genomic testing, TRECA's integration with sBeacon, enables small groups worldwide to easily share and discover variant data. If just 20% of these groups contribute, we could see a 6–7% improvement in diagnosis rates, resulting in 9–10 million additional life-changing rare disease diagnoses each year. Furthermore, the presence of Trusted Research Environments makes these rare disease research diagnoses reproducible and publication-ready.
What's next for TRECA
TRECA was built with a vision of one stop shop for clinicians and researchers. We will be working towards achieving this goal by continuously adding more shops for clinicians and researchers after taking feedback from real-world users. Here are the some of the requested features:
User-based cost tracker: While TRECA supports cost forecasting for individual user activities. We plan to enhance this feature to accumulate individuals compute usage and other costs over time.
Cater to infectious disease space: enhance TRECA to incorport our existing capability in pathogen genomics (PathsBeacon, a scalable query engine, and StrepiFun, an AI tool for understanding pathogen evolution).
FHIR to Beacon mapping: TRECA uses the beacon protocol to query genomic and phenotypic data together via sbeacon implementation. This allows any existing or custom ontology terms to be queried. FHIR and beacon mapping will further enhance this interoperability but careful consideration is needed to not lose context while mapping FHIR objects to beacon protocol.
Certified TRE: The group is actively working towards getting certification for TRE, i.e. as needed for Ukbiobank.
Dynamic Consent: We are working on cloud-based frameworks to capture and manage dynamic consent. We will incorporate this into TRECA in the future to manage the transition from clinic to research cohorts.
Federated Queries across TREs: TRECA enables federated querying within on instance, so across different projects, however we want to enable such queries and ultimately federated ML across different TRECA instances (i.e. cloud accounts).
Synthetic Data generation: We are working on Genomator, an algorithm for creating synthetic representation of data (i.e. digital twins). This aims to make the data more easily sharable as it makes data - including genomic data - unidentifiable by removing rare of private characteristics (i.e. SNPs) while maintaining potentially disease-characteristic allele frequencies at a population level.

