Exchanging pathogenic mutational information (e.g., SARS-CoV-2) in a fast and accessible way is important for tracking specific pathogen mutations (e.g., D614G). Comparing mutation frequencies can help identify a newly emerging outbreak cluster or elucidate the functional consequences of genomic evolution on the host, the pathogen, or the vaccine efficacy. Access to this type of mutational information is hence essential for developing effective treatments and vaccines, as well as mount a timely public health response. However, getting ahead of an emerging pandemic requires data to be shared in a fast, secure, and interoperable manner.
Genomic data exchange at scale
GA4GH’s (Global Alliance for Genomic Health) Beacon protocol is the accepted standard for sharing genomic and phenotypic information in human genomics. However, while the latest release (Version 2) allows the exchange of metadata, it is highly specific to the human genomics domain and not directly applicable to the pathogen domain.
We have adapted the Beacon protocol to build PathSBeacon, which is specialized on the exchange of infectious disease data and can be use to share viral mutation frequencies from distributed data sources. The adapted protocol ensures that data ownership is preserved, while enabling valuable insights to be generated from separate data sources. By creating a secure and accessible data-sharing platform, researchers and healthcare professionals can gain valuable insights from separate data sources, which can help identify trends and patterns that may be difficult to detect through individual data sets. PathSBeacon made a substantial contribution in the battle against the COVID-19 pandemic by enabling the exchange of critical information and insights to help combat the virus.
Beacon protocol for pathogenic data exchange
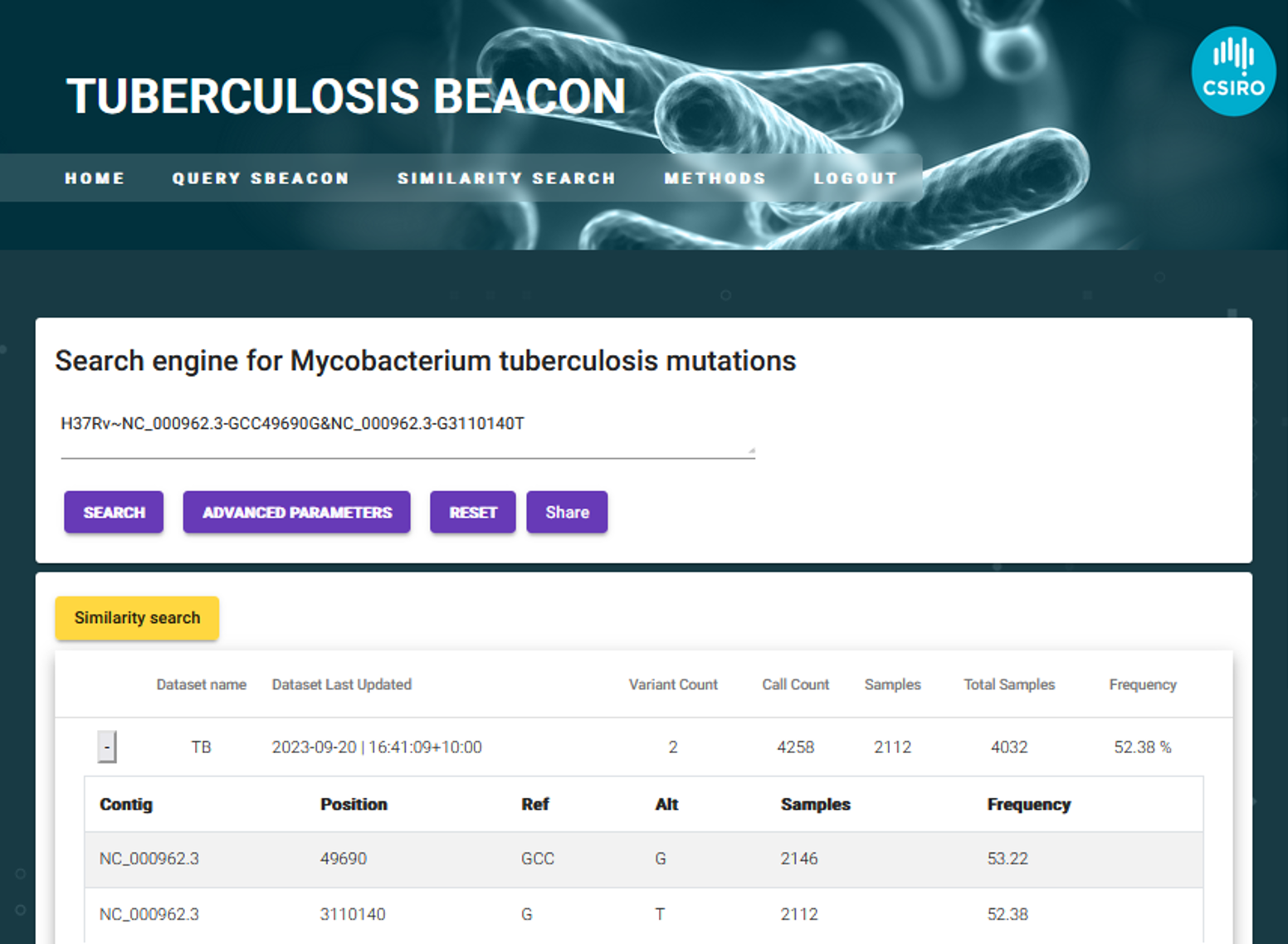
The following screenshot shows how queries can be performed in the PathSBeacon user interface. Popular searches can be defined in the quick search bar such as “D614G” or searching for all mutations in the Spike protein. PathsBeacon returns the genomic position of variant, change in allele, number of samples, SIFT score and frequency of the mutation, amongst other information.
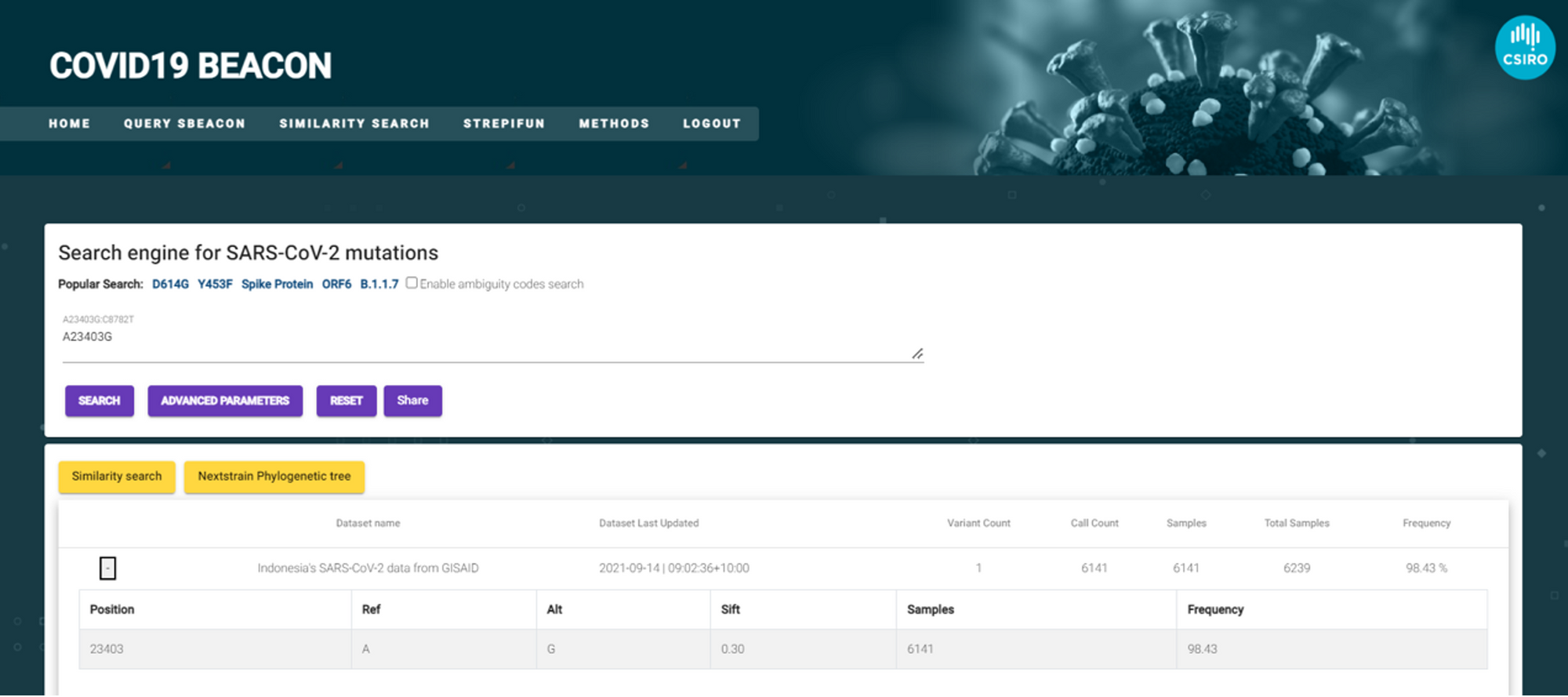
The following screenshot demonstrates the result returned which indicates the datasets discovered related to the search and basic information. These include number of variants, call count, total number of samples and the frequency. This information provides a snapshot of the beacon and the prevalence of a mutation of interest, providing a quick overview into each of the datasets.
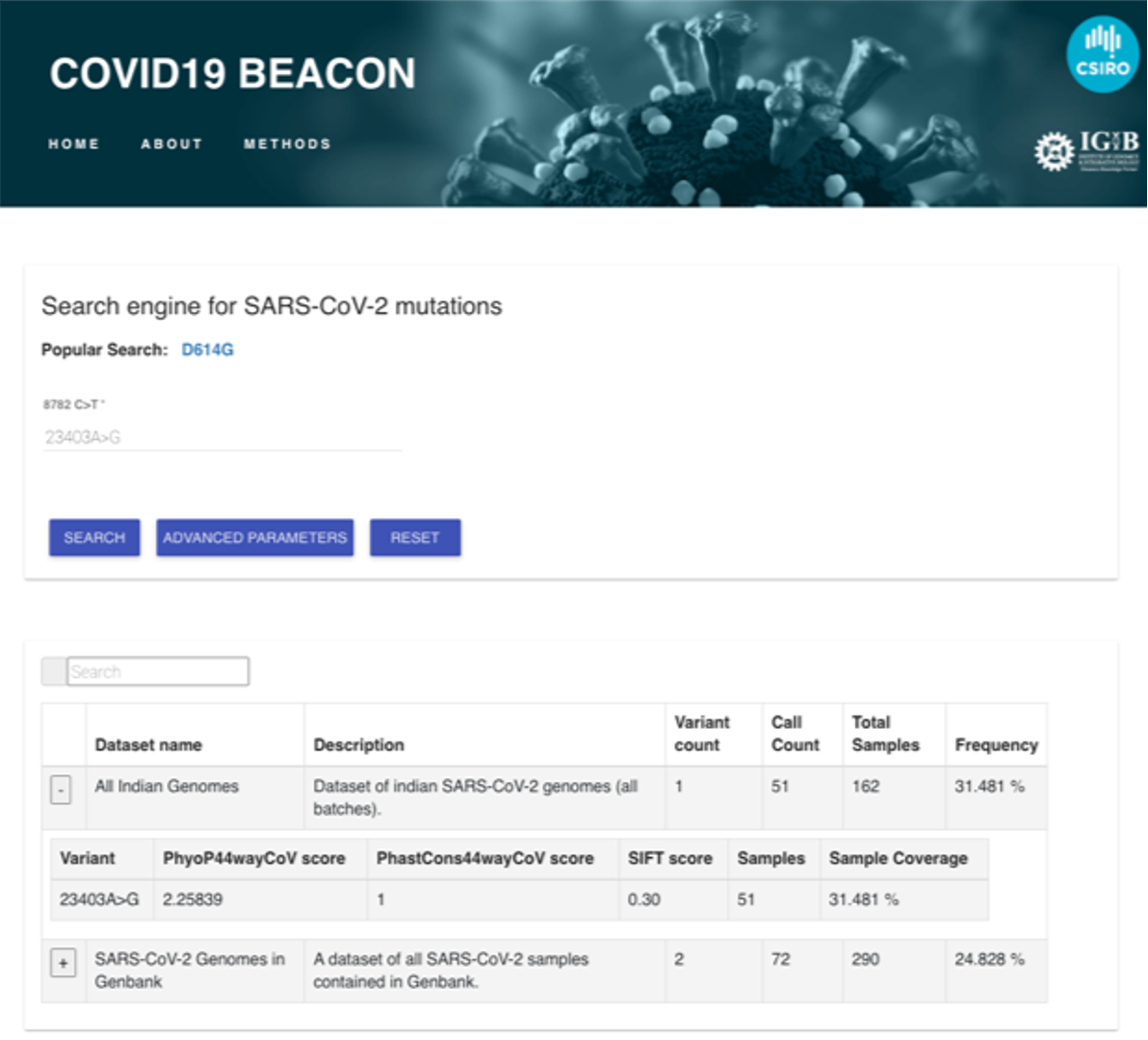
Unique value proposition of PathSBeacon
PathSBeacon is purpose build for tracking and querying infectious disease data, making global data exchange feasible and secure. It facilitates the exchange of observed allele frequencies in pathogen datasets alongside the following metadata information:
Sample collection date - timestamp of the collection moment
Location - geographical location of the sample's origin
State - location state (narrower geographic location)
Location:Sample collection date - combined location and date
State:Sample collection date - combined state and date
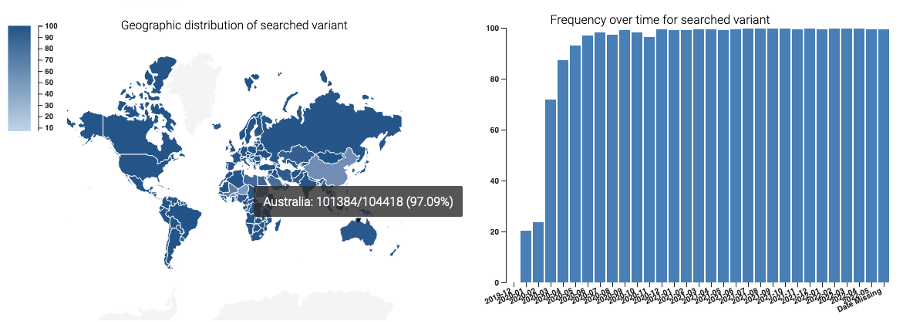
PathSBeacon provides advanced analytics over the onboarded datasets such as time series and geospatial statistics to illustrate the evolution and geographical distribution of pathogens.
PathSBeacon is pathogen agnostic and we developed alternate version of PathSBeacon to monitor other pathogens such as Tuberculosis and Gonorrhoea. Ongoing efforts are also invested in standardising the pathogen beacon requirements and the schema in collaboration with global bodies.
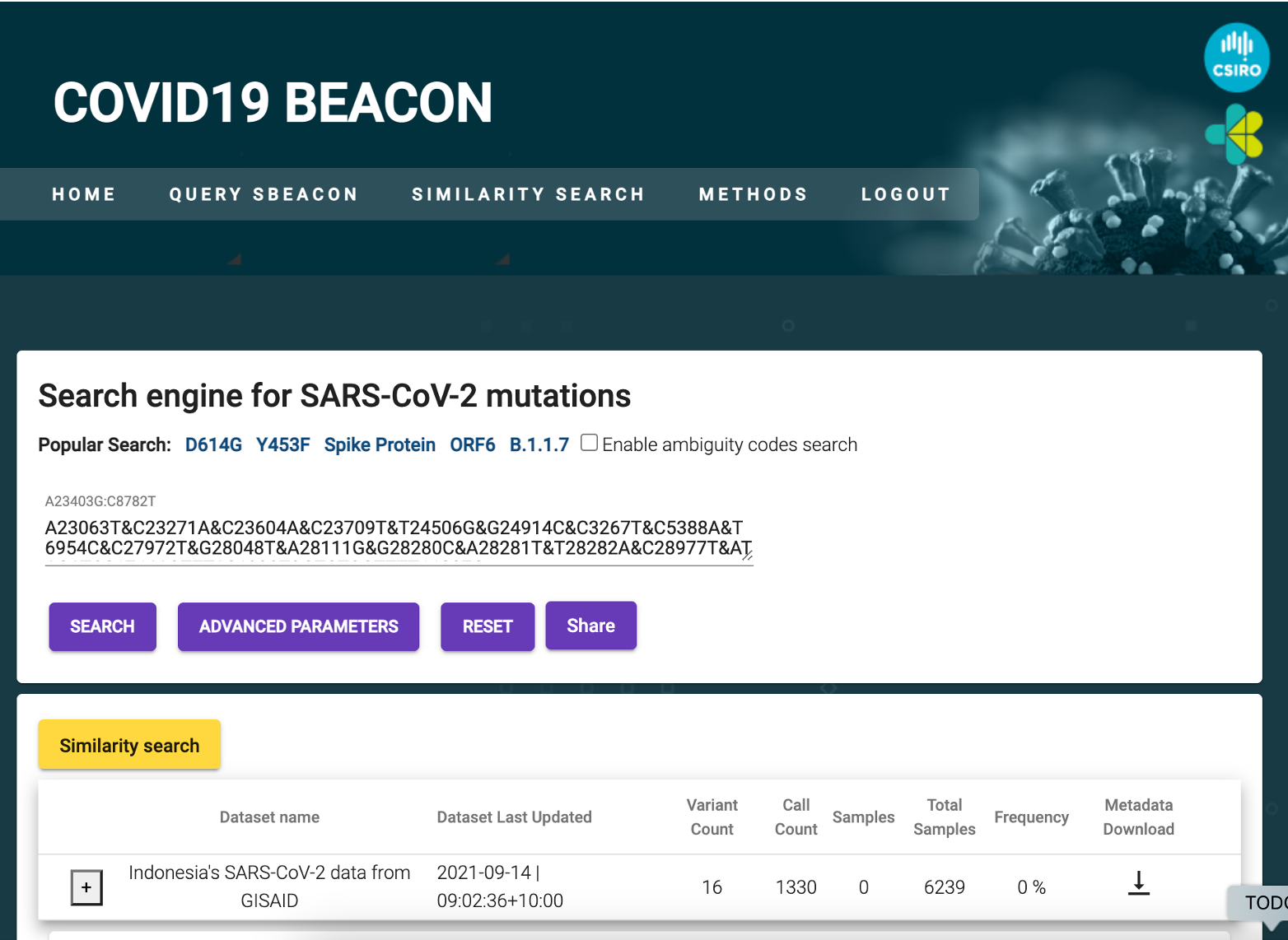

Global efforts in pathogenic data exchange
In March 2020 DNAstack announced that they also successfully adapted Beacon protocol to cater for RNA virus data resulting in the COVID-19 Beacon.
Comparison
Below we outline the outstanding feature of our Serverless COVID Beacon compared to the vanilla beacon protocol implementation. E.g. CSIRO's COVID Beacon, extends our Serverless Beacon, a cloud-native implementation of the Beacon protocol, which reduces resource consumption up to 500-fold, COVID-19 sBeacon is one of the most resource-efficient and functionally rich solutions for sharing viral variant information.
| Beacon | COVID-19 sBeacon | |
|---|---|---|
| variant frequency | ✔ | ✔ |
| IUPAC allowed | 𐄂 | ✔ |
| Range search | 𐄂 | ✔ |
| Clades | 𐄂 | ✔ |
| custom denominator | 𐄂 | ✔ |
Applications
We successfully deployed PathSBeacon with GSI lab, the second largest sequencing provider in Indonesia to track SARS-CoV-2 samples.
Read more - https://bioinformatics.csiro.au/blog/case-study-indonesias-pathsbeacon/
Pricing
- Custom implementation
- Bespoke solutions
- Product workshops
- Managed updates
- Full support

