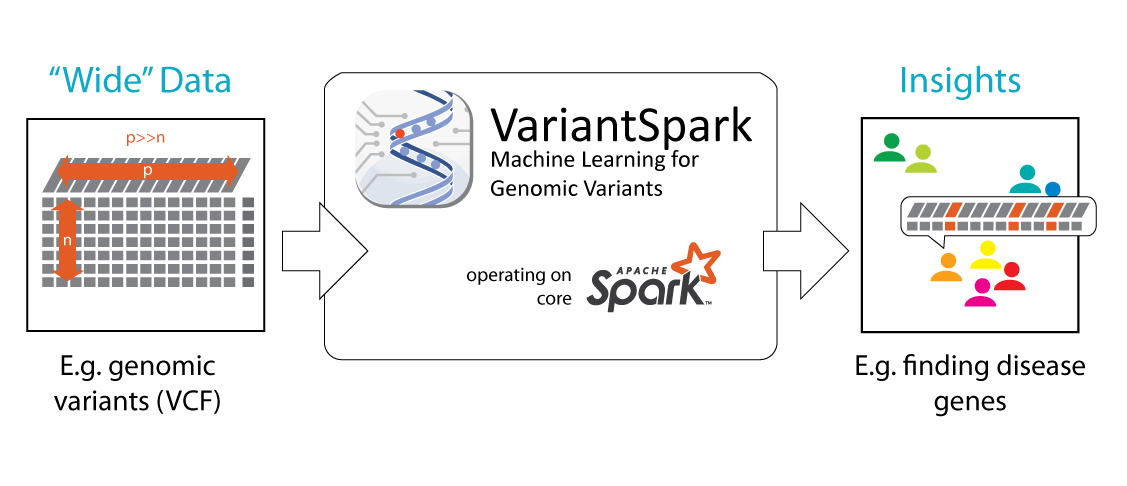
New approaches are needed to process modern datasets that are big and “wide” where millions of samples (rows) can have millions of information points each (columns, features).
We developed VariantSpark, a tailored Apache Spark-based machine learning framework that creates insights from high-dimensional data.
Value Proposition
Fast and sensitive end-to-end approach for high-dimensional data
VariantSpark is 90% faster than traditional compute frameworks and requires 80% fewer samples to detect statistical significant signal. The benefits of speed and higher sensitivity enables VariantSpark to open up the usage of advanced, efficient machine learning algorithms on high dimensional data.

Detect stronger predictive markers
VariantSpark’s machine learning method overcomes the limitation of traditional approaches that requires data to be eliminated or identifying only independent markers. Especially complex events are triggered by multiple contributing factors. VariantSpark is able to detect such sets of interacting features thereby identifying more accurate predictive markers.
Explainable ML
VariantSpark builds on the Random Forest Machine Learning method, which allows to interrogate the tree-based models and identify which features contributed in what proportion to the overall prediction outcome. We also provide a visualization engine that shows the interplay between features and their label association.
Awards
Pricing
- Custom implementation
- Bespoke solutions
- Product workshops
- Managed updates
- Full support
Do business with us
Let us be your innovation catalyst by helping you understand the health space, solve your pain-points and innovate to keep you ahead. Read more
Our Approach
Traditionally the columns (features) of large “raw” datasets needed to be scaled down to allow advanced analytics to take place. This process of eliminating information potentially biases the result and impacts on the sensitivity of the analysis. VariantSpark is different, it can deal with the raw data directly resulting in a more accurate output.
Variant Spark was tested on datasets with 3000 samples, each containing 80 Million features, in either unsupervised clustering approaches (e.g. k-means) or supervised applications (e.g. Random Forests) with target/truth values that are categorical (classification) or continuous (regression). It can cluster data according to required profiles or identify predictive markers of events in just 30 minutes.
By building a model on the full dataset, this allows, for the first time, to identify sets of markers that together have a stronger predictive power than the previously identified independent markers.
Application cases
- Disease gene detection where genomic loci can be detected that contribute jointly or individually to disease.
- Polygenic Risk Scores where genomic loci need to be prioritized for building a polygenic risk model, VariantSpark can select the top X features that individually or jointly influence outcome.
- Internet-of-Things where data extracted from IoT devices provides datasets with millions of observed features per sample, VariantSpark can provide companies with fresh insights into predictive markers about their customers and behaviours.
- Processing plant optimisation where millions of observations about the product property and supply chain can influence the quality of the resulting element, VariantSpark can identify specific components that contribute to optimal outcome.
- Churn rate optimisation where millions of factors can play into a customer's decision to churn. VariantSpark can predict which factors or combination of factors are most predictive of churn for your marketing and salesforce department to create an early warning sign indicator to prevent customer churn.
Case Studies
Disease gene detection with the Hipster data on Databricks
VariantSpark is developed for 'big' (many samples, n) and 'wide' (large feature vector per sample, p) data. It was tested on datasets with n=3000 samples each containing p=80Million features in either unsupervised clustering approaches (e.g. k-means) or supervised applications with target/truth values that are categorical (classification) or continuous (regression). Though VariantSpark was originally developed for genomic variant data, where p={0,1,2}, it can cater for every feature-based dataset, e.g. methylation, transcription, non-biological applications.
Accessibility
Click on the icons to see all our activities with these vendors.
Collaboration Partners
Getting Started
To help you get started we've put together a couple of videos.
1) Using VariantSpark on AWS Marketplace
Video coming soon. Meanwhile read Lynn Langit's Medium article.
2) Using VariantSpark on Databricks
3) Spinning up VariantSpark on AWS using cloudformation